Image Processing
Pipeline description
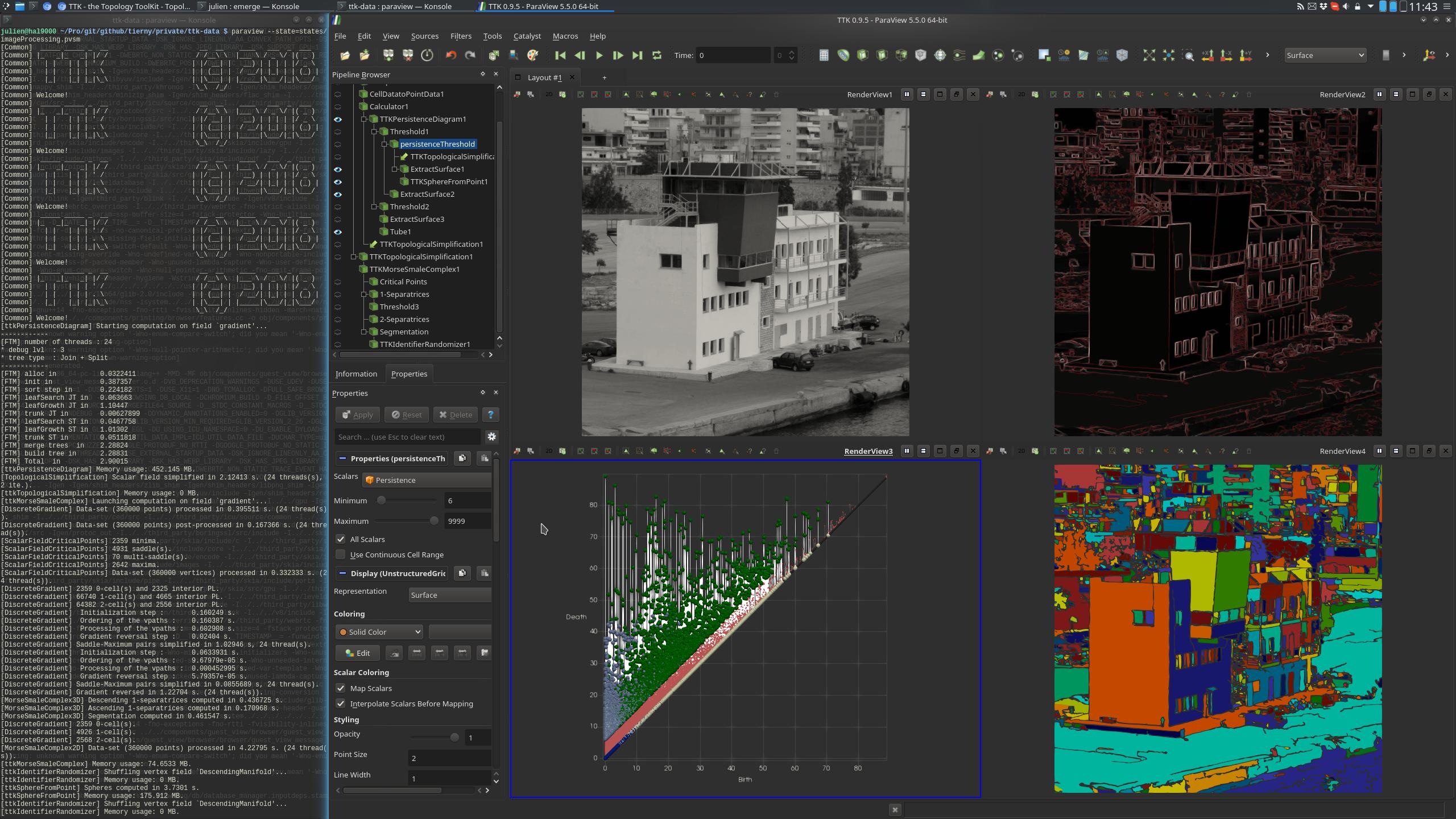
This example processes a grayscale image (top left view on the
above screenshot) to generate a segmentation. We will construct the
segmentation from the image gradient.
First, the image is loaded from disk. The gradient is computed with
ParaView's ComputeDerivatives or Gradient filters. Since TTK only
works on scalar field, a Calculator is used to compute the gradient
magnitude.
From the gradient magnitude, a simplification step involving
PersistenceDiagram
(bottom left view) and
TopologicalSimplification
helps removing the noise in the gradient (top right view).
To segment the image, we use the
MorseSmaleComplex
filter. Since in the input image the objects correspond to low values
in the gradient and their edges to high values, we are interested in
the DescendingManifold scalar field of the Segmentation output, whose
cells represent regions of low scalar field values.
The
IdentifierRandomizer
filter is eventually used in order to color neighbor cells with a
distinct color (bottom right view on the above screenshot).
ParaView
To reproduce the above screenshot, go to your
ttk-data directory
and enter the following command:
paraview states/imageProcessing.pvsm
Python code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61 | #!/usr/bin/env python
from paraview.simple import *
naturalImagepng = PNGSeriesReader(FileNames=["naturalImage.png"])
# create a new 'Compute Derivatives'
computeDerivatives1 = ComputeDerivatives(Input=naturalImagepng)
computeDerivatives1.Scalars = ["POINTS", "PNGImage"]
# create a new 'Cell Data to Point Data'
cellDatatoPointData1 = CellDatatoPointData(Input=computeDerivatives1)
# create a new 'Calculator'
calculator1 = Calculator(Input=cellDatatoPointData1)
calculator1.ResultArrayName = "gradient"
calculator1.Function = "mag(ScalarGradient)"
# create a new 'TTK PersistenceDiagram'
tTKPersistenceDiagram1 = TTKPersistenceDiagram(Input=calculator1)
tTKPersistenceDiagram1.ScalarField = ["POINTS", "gradient"]
# create a new 'Threshold'
threshold1 = Threshold(Input=tTKPersistenceDiagram1)
threshold1.Scalars = ["CELLS", "PairIdentifier"]
threshold1.ThresholdMethod = "Between"
threshold1.LowerThreshold = 0.0
threshold1.UpperThreshold = 999999999
# create a new 'Threshold'
persistenceThreshold = Threshold(Input=threshold1)
persistenceThreshold.Scalars = ["CELLS", "Persistence"]
persistenceThreshold.ThresholdMethod = "Between"
persistenceThreshold.LowerThreshold = 6.0
persistenceThreshold.UpperThreshold = 999999999
# create a new 'TTK TopologicalSimplification'
tTKTopologicalSimplification1 = TTKTopologicalSimplification(
Domain=calculator1,
Constraints=persistenceThreshold,
)
tTKTopologicalSimplification1.ScalarField = ["POINTS", "gradient"]
# create a new 'TTK MorseSmaleComplex'
tTKMorseSmaleComplex1 = TTKMorseSmaleComplex(Input=tTKTopologicalSimplification1)
tTKMorseSmaleComplex1.ScalarField = ["POINTS", "gradient"]
# create a new 'Threshold'
threshold3 = Threshold(Input=OutputPort(tTKMorseSmaleComplex1, 1))
threshold3.Scalars = ["CELLS", "SeparatrixType"]
threshold3.ThresholdMethod = "Between"
threshold3.LowerThreshold = 1.0
threshold3.UpperThreshold = 1.0
# create a new 'TTK IdentifierRandomizer'
tTKIdentifierRandomizer1 = TTKIdentifierRandomizer(
Input=OutputPort(tTKMorseSmaleComplex1, 3),
)
tTKIdentifierRandomizer1.ScalarField = ["POINTS", "DescendingManifold"]
SaveData("Segmentation.vti", tTKIdentifierRandomizer1)
|
To run the above Python script, go to your ttk-data directory and enter the following command:
pvpython python/imageProcessing.py
Outputs
Segmentation.vti: the image segmentation output.
C++/Python API
IdentifierRandomizer
MorseSmaleComplex
PersistenceDiagram
TopologicalSimplification
