Manifold Check
Pipeline description
This example loads three different hexahedral geometry files from disk.
In a pre-processing, each geometry is tetrahedralized, which is used as input data.
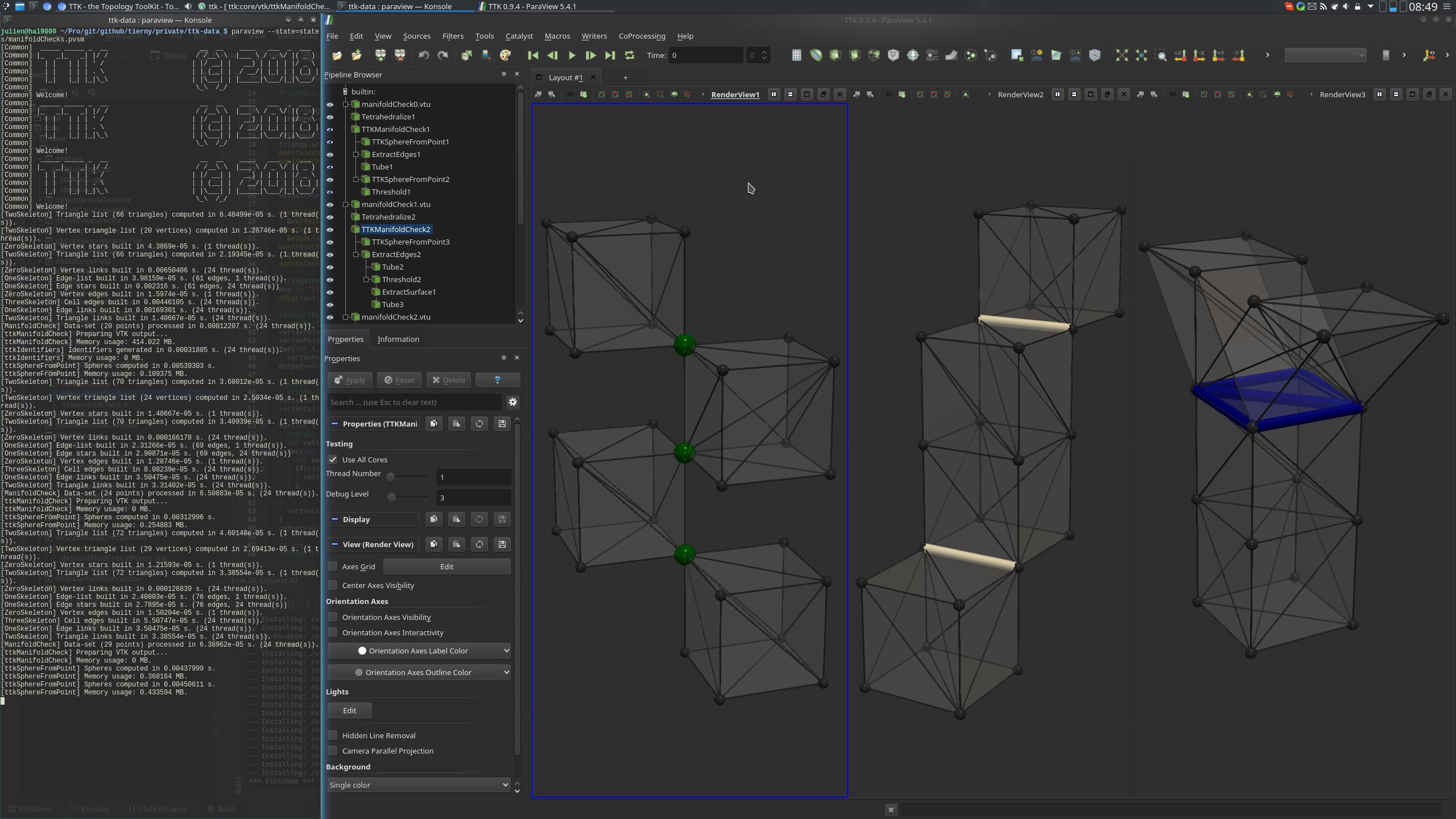
On each of the three geometries, ManifoldCheck is executed. This filters adds link numbers to vertices and cells, which can be used to detect and extract non-manifold vertices (left), edges (middle), and faces (right).
ParaView
To reproduce the above screenshot, go to your ttk-data directory and enter the following command:
paraview states/manifoldChecks.pvsm
Python code
Non-manifold Vertices
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30 | #!/usr/bin/env python
from paraview.simple import *
# create a new 'XML Unstructured Grid Reader'
manifoldCheck0vtu = XMLUnstructuredGridReader(FileName=["manifoldCheck0.vtu"])
# create a new 'Tetrahedralize'
tetrahedralize1 = Tetrahedralize(Input=manifoldCheck0vtu)
# create a new 'TTK ManifoldCheck'
tTKManifoldCheck1 = TTKManifoldCheck(Input=tetrahedralize1)
# create a new 'Mask Points'
maskPoints1 = MaskPoints(Input=tTKManifoldCheck1)
maskPoints1.OnRatio = 1
maskPoints1.MaximumNumberofPoints = 1000
maskPoints1.GenerateVertices = 1
maskPoints1.SingleVertexPerCell = 1
# create a new 'Threshold'
# this extracts non-manifold vertices
threshold1 = Threshold(Input=maskPoints1)
threshold1.Scalars = ["POINTS", "VertexLinkComponentNumber"]
threshold1.ThresholdMethod = "Between"
threshold1.LowerThreshold = 2.0
threshold1.UpperThreshold = 2.0
# save the output
SaveData("manifoldCheck0_check.vtu", tTKManifoldCheck1)
SaveData("manifoldCheck0_non_manifold.vtu", threshold1)
|
To run the above Python script, go to your ttk-data directory and enter the following command:
pvpython python/manifoldCheck0.py
Non-manifold Edges
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26 | #!/usr/bin/env python
from paraview.simple import *
# create a new 'XML Unstructured Grid Reader'
manifoldCheck1vtu = XMLUnstructuredGridReader(FileName=["manifoldCheck1.vtu"])
# create a new 'Tetrahedralize'
tetrahedralize2 = Tetrahedralize(Input=manifoldCheck1vtu)
# create a new 'TTK ManifoldCheck'
tTKManifoldCheck2 = TTKManifoldCheck(Input=tetrahedralize2)
# create a new 'Extract Edges'
extractEdges2 = ExtractEdges(Input=tTKManifoldCheck2)
# create a new 'Threshold'
# this extracts non-manifold edges
threshold2 = Threshold(Input=extractEdges2)
threshold2.Scalars = ["POINTS", "EdgeLinkComponentNumber"]
threshold2.ThresholdMethod = "Between"
threshold2.LowerThreshold = 2.0
threshold2.UpperThreshold = 2.0
# save the output
SaveData("manifoldCheck1_check.vtu", tTKManifoldCheck2)
SaveData("manifoldCheck1_non_manifold.vtu", threshold2)
|
To run the above Python script, go to your ttk-data directory and enter the following command:
pvpython python/manifoldCheck1.py
Non-manifold Faces
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51 | #!/usr/bin/env python
from paraview.simple import *
# create a new 'XML Unstructured Grid Reader'
manifoldCheck2vtu = XMLUnstructuredGridReader(FileName=["manifoldCheck2.vtu"])
# create a new 'Tetrahedralize'
tetrahedralize3 = Tetrahedralize(
registrationName="Tetrahedralize3", Input=manifoldCheck2vtu
)
# create a new 'TTK ManifoldCheck'
tTKManifoldCheck3 = TTKManifoldCheck(
registrationName="TTKManifoldCheck3", Input=tetrahedralize3
)
# create a new 'Threshold'
# this extracts tetrahedra that contain non-manifold faces
threshold3 = Threshold(registrationName="Threshold3", Input=tTKManifoldCheck3)
threshold3.Scalars = ["CELLS", "TriangleLinkComponentNumber"]
threshold3.ThresholdMethod = "Between"
threshold3.LowerThreshold = 3.0
threshold3.UpperThreshold = 3.0
# create a new 'Generate Ids'
generateIds1 = GenerateIds(registrationName="GenerateIds1", Input=threshold3)
generateIds1.PointIdsArrayName = "VertexIdentifiers"
generateIds1.CellIdsArrayName = "CellIdentifiers"
# create a new 'Threshold'
# select two of the tetrahedra
threshold4 = Threshold(registrationName="Threshold4", Input=generateIds1)
threshold4.Scalars = ["CELLS", "CellIdentifiers"]
threshold4.ThresholdMethod = "Between"
threshold4.LowerThreshold = 0.0
threshold4.UpperThreshold = 1.0
# create a new 'Extract Surface'
extractSurface2 = ExtractSurface(registrationName="ExtractSurface2", Input=threshold4)
# create a new 'Threshold'
# this extracts non-manifold faces
threshold5 = Threshold(registrationName="Threshold5", Input=extractSurface2)
threshold5.Scalars = ["POINTS", "TriangleLinkComponentNumber"]
threshold5.ThresholdMethod = "Between"
threshold5.LowerThreshold = 3.0
threshold5.UpperThreshold = 3.0
# save the output
SaveData("manifoldCheck2_check.vtu", tTKManifoldCheck3)
SaveData("manifoldCheck2_non_manifold.vtu", threshold5)
|
To run the above Python script, go to your ttk-data directory and enter the following command:
pvpython python/manifoldCheck2.py
Outputs
manifoldCheck0_check.vtu, manifoldCheck1_check.vtu, manifoldCheck2_check.vtu: tetrhedralized geometry with link numbersmanifoldCheck0_non_manifold.vtu: non-manifold vertices in manifoldCheck0.vtumanifoldCheck1_non_manifold.vtu: non-manifold edges in manifoldCheck1.vtumanifoldCheck2_non_manifold.vtu: non-manifold faces in manifoldCheck2.vtu
C++/Python API
ManifoldCheck
