Morse-Smale Segmentation AT
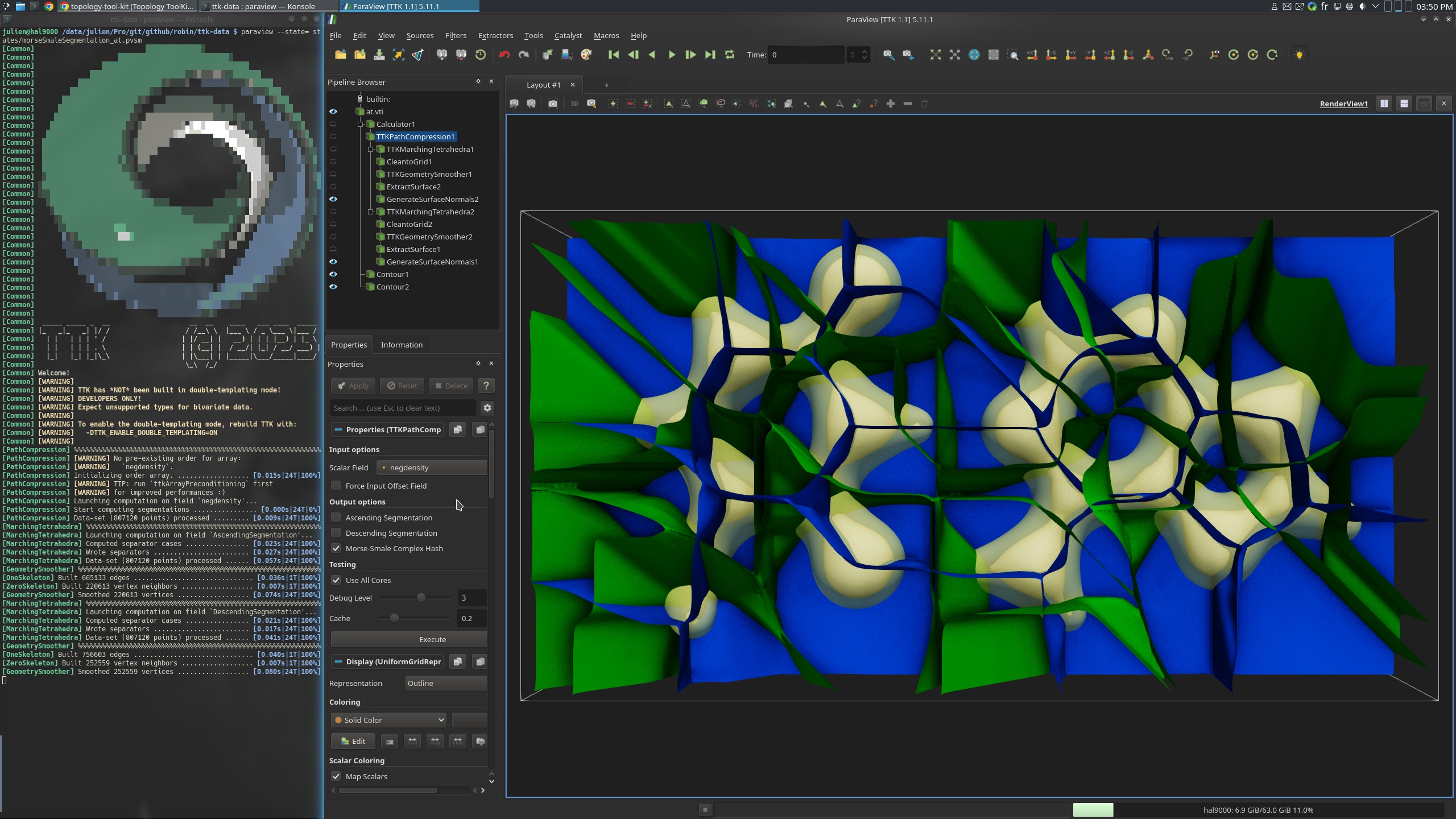
Pipeline description
This example computes the ascending and descending Morse segmentations from an electronic density of the Adenine-Thymine molecular complex.
The descending segmentation separators (red) represent the influence areas of maxima, giving each atom its own pocket since they are the most dense points in the dataset. The ascending segmentation separators (blue) represent the influence area of minima, providing the interactions (covalent and non-covalent) between the atoms in this example.
The segmentations are computed using PathCompression.
The separating geometry is generated using MarchingTetrahedra
The python script simply computes the segmentation and saves the separating
surfaces as .vtu files.
ParaView
To reproduce the above screenshot, go to your ttk-data directory and enter the following command:
paraview states/morseSmaleSegmentation_at.pvsm
Python code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54 | #!/usr/bin/env python
from paraview.simple import *
# create a new 'XML Image Data Reader'
atvti = XMLImageDataReader(FileName=["at.vti"])
# create a new 'Calculator'
calculator1 = Calculator(registrationName="Calculator1", Input=atvti)
calculator1.ResultArrayName = "negdensity"
calculator1.Function = "-density"
# create a new 'TTK PathCompression'
tTKPathCompression1 = TTKPathCompression(
registrationName="TTKPathCompression1", Input=calculator1
)
tTKPathCompression1.ScalarField = ["POINTS", "negdensity"]
# create a new 'TTK MarchingTetrahedra'
tTKMarchingTetrahedra1 = TTKMarchingTetrahedra(
registrationName="TTKMarchingTetrahedra1", Input=tTKPathCompression1
)
tTKMarchingTetrahedra1.ScalarField = ["POINTS", "negdensity_DescendingManifold"]
# create a new 'Clean to Grid'
cleantoGrid1 = CleantoGrid(
registrationName="CleantoGrid1", Input=tTKMarchingTetrahedra1
)
# create a new 'TTK GeometrySmoother'
tTKGeometrySmoother1 = TTKGeometrySmoother(
registrationName="TTKGeometrySmoother1", Input=cleantoGrid1
)
tTKGeometrySmoother1.IterationNumber = 20
# create a new 'TTK MarchingTetrahedra'
tTKMarchingTetrahedra2 = TTKMarchingTetrahedra(
registrationName="TTKMarchingTetrahedra2", Input=tTKPathCompression1
)
tTKMarchingTetrahedra2.ScalarField = ["POINTS", "negdensity_AscendingManifold"]
# create a new 'Clean to Grid'
cleantoGrid2 = CleantoGrid(
registrationName="CleantoGrid2", Input=tTKMarchingTetrahedra2
)
# create a new 'TTK GeometrySmoother'
tTKGeometrySmoother2 = TTKGeometrySmoother(
registrationName="TTKGeometrySmoother2", Input=cleantoGrid2
)
tTKGeometrySmoother2.IterationNumber = 20
SaveData("descendingSeparatorAt.vtu", tTKGeometrySmoother1)
SaveData("ascendingSeparatorAt.vtu", tTKGeometrySmoother2)
|
To run the above Python script, go to your ttk-data directory and enter the following command:
pvpython python/morseSmaleSegmentation_at.py
- at.vti: A molecular dataset: a three-dimensional regular grid with 1 scalar field, the electronic density in the Adenine Thymine complex.
Outputs
ascendingSeparatorAt.vtu: the ascending separator geometry.descendingSeparatorAt.vtu: the descending separator geometry.
C++/Python API
GeometrySmoother
MarchingTetrahedra
PathCompression
