Persistent Generators Periodic Picture
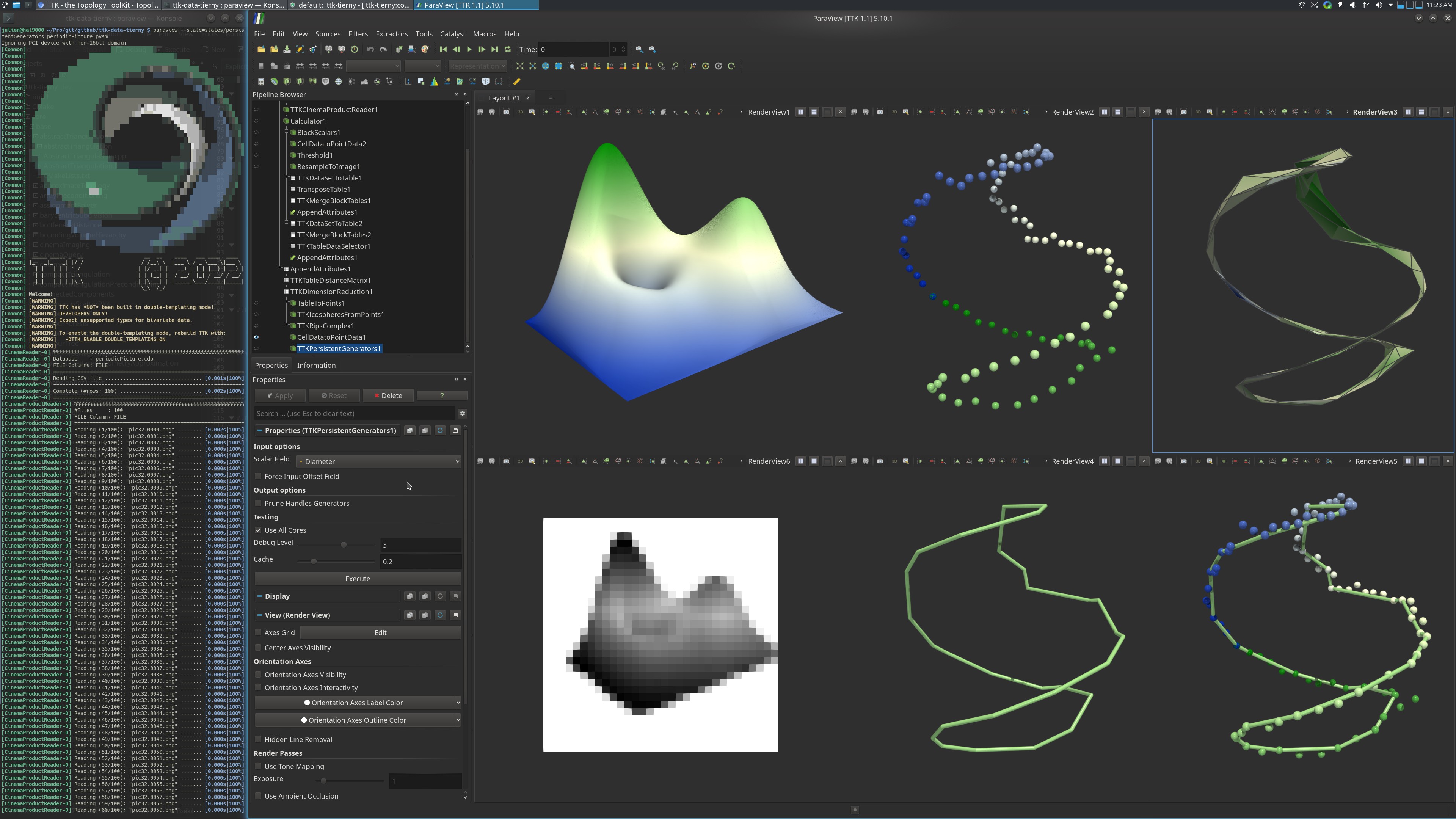
Pipeline description
This example shows the detection of a circular pattern in high-dimensional data.
100 greyscale pictures of resolution 32x32 of a syntethic terrain were taken in ParaView, from 100 viewpoints arranged along a circle, and stored in a cinema database.
The ParaView example starts by loading the database. The set of pictures and the corresponding viewpoints can be navigated using the time controls of ParaView.
This set of images can be interpreted as 100 points in R^1024.
The distance matrix between these points is computed using TableDistanceMatrix.
These points are then embedded in 3D using MDS, with DimensionReduction.
At this point, the 2-dimensional Rips complex is computed with RipsComplex from the high-dimensional point cloud, and shown in 3D via MDS projection.
Finally the infinitely persistent 1-cycle is extracted from the Rips complex with PersistentGenerators, on the diameter function.
This cycle captures the circular pattern synthetically injected in the data.
The python script saves the resulting point cloud, Rips complex and output cycle as vtk files.
ParaView
To reproduce the above screenshot, go to your ttk-data directory and enter the following command:
paraview states/persistentGenerators_periodicPicture.pvsm
Python code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64 | #!/usr/bin/env python
from paraview.simple import *
# create a new 'TTK CinemaReader'
tTKCinemaReader1 = TTKCinemaReader(DatabasePath="periodicPicture.cdb")
# create a new 'TTK CinemaProductReader'
tTKCinemaProductReader1 = TTKCinemaProductReader(Input=tTKCinemaReader1)
# create a new 'Calculator'
calculator1 = Calculator(Input=tTKCinemaProductReader1)
calculator1.Function = "PNGImage_X"
# create a new 'TTK DataSetToTable'
tTKDataSetToTable1 = TTKDataSetToTable(Input=calculator1)
tTKDataSetToTable1.DataAssociation = "Point"
# create a new 'Transpose Table'
transposeTable1 = TransposeTable(Input=tTKDataSetToTable1)
transposeTable1.VariablesofInterest = ["Result"]
# create a new 'TTK MergeBlockTables'
tTKMergeBlockTables1 = TTKMergeBlockTables(Input=transposeTable1)
# create a new 'TTK TableDistanceMatrix'
tTKTableDistanceMatrix1 = TTKTableDistanceMatrix(Input=tTKMergeBlockTables1)
tTKTableDistanceMatrix1.SelectFieldswithaRegexp = 1
tTKTableDistanceMatrix1.Regexp = "[01].*"
# create a new 'TTK DimensionReduction'
tTKDimensionReduction1 = TTKDimensionReduction(
Input=tTKTableDistanceMatrix1, ModulePath="default"
)
tTKDimensionReduction1.SelectFieldswithaRegexp = 1
tTKDimensionReduction1.Regexp = "Dist.*"
tTKDimensionReduction1.Components = 3
tTKDimensionReduction1.InputIsaDistanceMatrix = 1
tTKDimensionReduction1.UseAllCores = 0
# create a new 'TTK RipsComplex'
tTKRipsComplex1 = TTKRipsComplex(Input=tTKDimensionReduction1)
tTKRipsComplex1.SelectFieldswithaRegexp = 1
tTKRipsComplex1.Regexp = "Dist.*"
tTKRipsComplex1.Diameterepsilon = 1500.0
# create a new 'Cell Data to Point Data'
cellDatatoPointData1 = CellDatatoPointData(Input=tTKRipsComplex1)
cellDatatoPointData1.CellDataArraytoprocess = ["Diameter"]
# create a new 'TTK PersistentGenerators'
tTKPersistentGenerators1 = TTKPersistentGenerators(Input=cellDatatoPointData1)
tTKPersistentGenerators1.ScalarField = ["POINTS", "Diameter"]
# create a new 'Table To Points'
tableToPoints1 = TableToPoints(Input=tTKDimensionReduction1)
tableToPoints1.XColumn = "Component_0"
tableToPoints1.YColumn = "Component_1"
tableToPoints1.ZColumn = "Component_2"
tableToPoints1.KeepAllDataArrays = 1
SaveData("PersistentGeneratorsPeriodicPicture_cycle.vtp", tTKPersistentGenerators1)
SaveData("PersistentGeneratorsPeriodicPicture_points.vtp", tableToPoints1)
SaveData("PersistentGeneratorsPeriodicPicture_ripsComplex.vtu", tTKRipsComplex1)
|
To run the above Python script, go to your ttk-data directory and enter the following command:
pvpython python/persistentGenerators_periodicPicture.py
- periodicPicture.cdb: A cinema database containing all 100 pictures, with metadata (the associated camera angle).
Outputs
PersistentGeneratorsPeriodicPicture_points.vtp: the high-dimensionnal point cloud embedded in 3D.PersistentGeneratorsPeriodicPicture_ripsComplex.vtu: the 2-dimensional Rips complex of the point cloud.PersistentGeneratorsPeriodicPicture_cycle.vtp: the infinitely persistent 1-cycle of the Rips complex.
C++/Python API
CinemaReader
CinemaProductReader
DataSetToTable
DimensionReduction
MergeBlockTables
PersistentGenerators
RipsComplex
TableDistanceMatrix
