Merge Tree Clustering
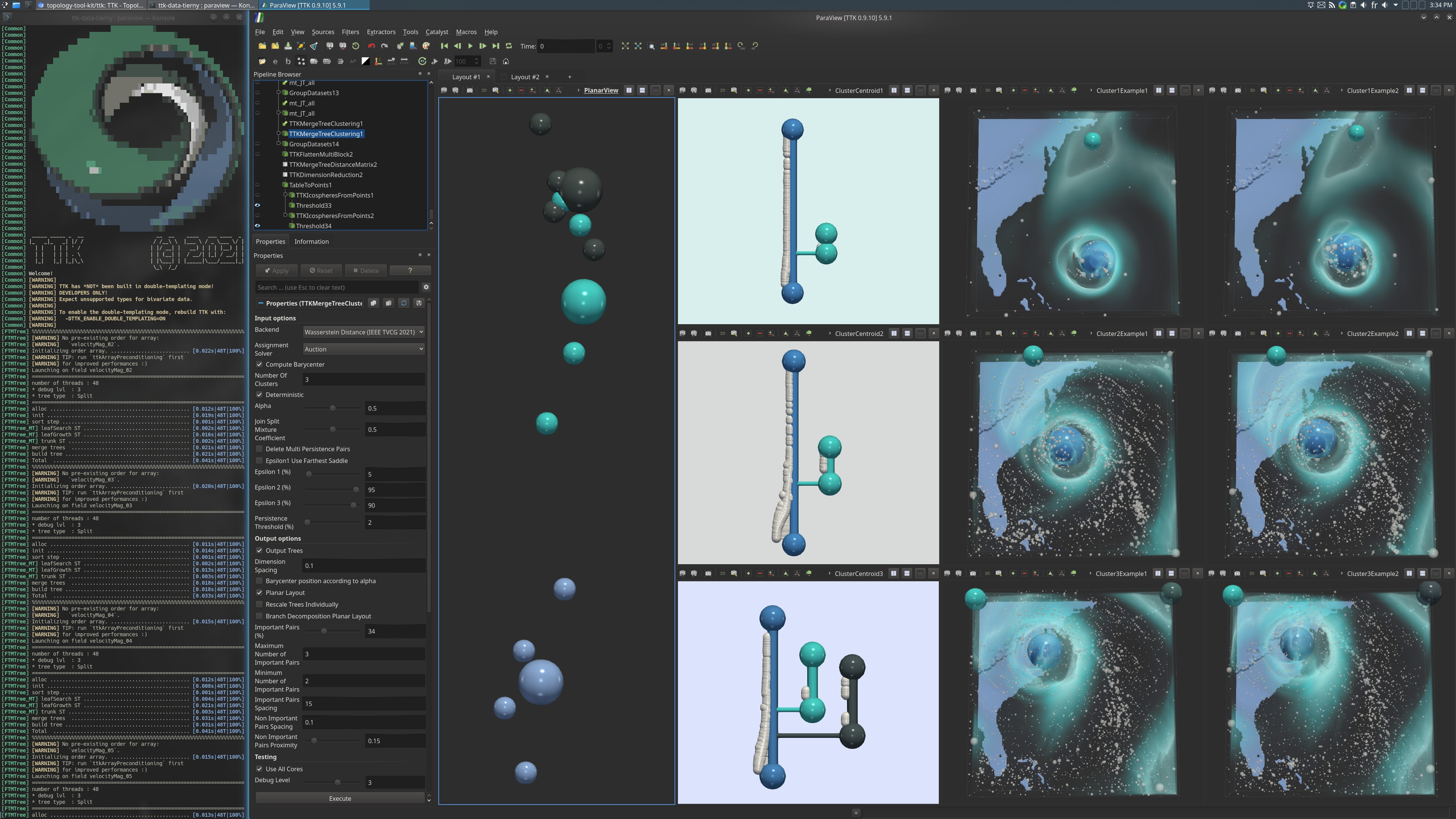
Pipeline description
This example first loads an ensemble of scalar fields inside a cinema database from disk.
Then, the MergeTree is computed on each scalar field for the Join Tree and the Split Tree.
All these trees are passed to MergeTreeClustering to compute a clustering in the metric space of merge trees. Each input is considered as a tuple consisting of the Join Tree and the Split Tree of the corresponding scalar field. Each centroid is also a tuple of this kind and a distance between two tuples is the distance between their Join Tree plus the distance between their Split Trees.
Then, a distance matrix is computed with MergeTreeDistanceMatrix with the input trees and the 3 centroids.
This distance matrix is used as input of DimensionReduction to compute a MultiDimensional Scaling (MDS), performing a dimensionality reduction in 2D respecting the most the input distance matrix.
In terms of visualisation, the MDS result is visualized and colored by clustering assignment. The split trees centroids are visualized with a planar layout and also some fields of each cluster.
In the second layout, the star clustering is visualized, consisting of the input split trees grouped by cluster, with the centroid of the cluster in the middle.
The python script computes the MDS and saves the resulting 2D points (for input trees and centroids).
ParaView
To reproduce the above screenshot, go to your ttk-data directory and enter the following command:
araview states/mergeTreeClustering.pvsm
Python code
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
98
99
100
101
102
103
104
105
106
107
108
109
110
111
112
113
114
115
116
117
118
119
120
121
122
123
124
125
126
127
128
129
130
131
132
133
134
135
136
137
138
139
140 | #!/usr/bin/env python
from paraview.simple import *
# create a new 'TTK CinemaReader'
tTKCinemaReader1 = TTKCinemaReader(DatabasePath="./Isabel.cdb")
# create a new 'TTK CinemaProductReader'
tTKCinemaProductReader1 = TTKCinemaProductReader(Input=tTKCinemaReader1)
tTKCinemaProductReader1.AddFieldDataRecursively = 1
# create a new 'TTK Merge and Contour Tree'
tTKMergeandContourTreeFTM26 = TTKMergeTree(Input=tTKCinemaProductReader1)
tTKMergeandContourTreeFTM26.ScalarField = ["POINTS", "velocityMag"]
tTKMergeandContourTreeFTM26.TreeType = "Join Tree"
# create a new 'Group Datasets'
mt_JT_all = GroupDatasets(
Input=[
tTKMergeandContourTreeFTM26,
OutputPort(tTKMergeandContourTreeFTM26, 1),
OutputPort(tTKMergeandContourTreeFTM26, 2),
]
)
# create a new 'TTK Merge and Contour Tree'
tTKMergeandContourTreeFTM25 = TTKMergeTree(Input=tTKCinemaProductReader1)
tTKMergeandContourTreeFTM25.ScalarField = ["POINTS", "velocityMag"]
tTKMergeandContourTreeFTM25.TreeType = "Split Tree"
# create a new 'Group Datasets'
mT_all = GroupDatasets(
Input=[
tTKMergeandContourTreeFTM25,
OutputPort(tTKMergeandContourTreeFTM25, 1),
OutputPort(tTKMergeandContourTreeFTM25, 2),
]
)
# create a new 'TTK MergeTreeClustering'
tTKMergeTreeClustering1 = TTKMergeTreeClustering(
Input=mT_all, OptionalInputclustering=mt_JT_all
)
tTKMergeTreeClustering1.ComputeBarycenter = 1
tTKMergeTreeClustering1.NumberOfClusters = 3
tTKMergeTreeClustering1.Deterministic = 1
tTKMergeTreeClustering1.DimensionSpacing = 0.1
tTKMergeTreeClustering1.PersistenceThreshold = 2.0
tTKMergeTreeClustering1.ImportantPairs = 34.0
tTKMergeTreeClustering1.MaximumNumberofImportantPairs = 3
tTKMergeTreeClustering1.MinimumNumberofImportantPairs = 2
tTKMergeTreeClustering1.ImportantPairsSpacing = 15.0
tTKMergeTreeClustering1.NonImportantPairsProximity = 0.15
# create a new 'Extract Block'
nodes = ExtractBlock(Input=OutputPort(tTKMergeTreeClustering1, 1))
nodes.Selectors = ["/Root/Block0"]
# create a new 'Extract Block'
nodes_1 = ExtractBlock(Input=tTKMergeTreeClustering1)
nodes_1.Selectors = ["/Root/Block0"]
# create a new 'Extract Block'
arcs = ExtractBlock(Input=OutputPort(tTKMergeTreeClustering1, 1))
arcs.Selectors = ["/Root/Block1"]
# create a new 'Extract Block'
arcs_1 = ExtractBlock(Input=tTKMergeTreeClustering1)
arcs_1.Selectors = ["/Root/Block1"]
# create a new 'TTK BlockAggregator'
tTKBlockAggregator1 = TTKBlockAggregator(
registrationName="TTKBlockAggregator1", Input=[nodes_1, nodes]
)
# create a new 'TTK FlattenMultiBlock'
tTKFlattenMultiBlock1 = TTKFlattenMultiBlock(
registrationName="TTKFlattenMultiBlock1", Input=tTKBlockAggregator1
)
# create a new 'TTK BlockAggregator'
tTKBlockAggregator2 = TTKBlockAggregator(
registrationName="TTKBlockAggregator2", Input=[arcs_1, arcs]
)
# create a new 'TTK FlattenMultiBlock'
tTKFlattenMultiBlock3 = TTKFlattenMultiBlock(
registrationName="TTKFlattenMultiBlock3", Input=tTKBlockAggregator2
)
# create a new 'TTK BlockAggregator'
tTKBlockAggregator3 = TTKBlockAggregator(
registrationName="TTKBlockAggregator3",
Input=[tTKFlattenMultiBlock1, tTKFlattenMultiBlock3],
)
tTKBlockAggregator3.FlattenInput = 0
# create a new 'TTK MergeTreeDistanceMatrix'
tTKMergeTreeDistanceMatrix2 = TTKMergeTreeDistanceMatrix(Input=tTKBlockAggregator3)
tTKMergeTreeDistanceMatrix2.PersistenceThreshold = 2.0
# create a new 'TTK DimensionReduction'
tTKDimensionReduction2 = TTKDimensionReduction(
Input=tTKMergeTreeDistanceMatrix2, ModulePath="default"
)
tTKDimensionReduction2.SelectFieldswithaRegexp = 1
tTKDimensionReduction2.Regexp = "Tree[0-9]+"
tTKDimensionReduction2.InputIsaDistanceMatrix = 1
tTKDimensionReduction2.UseAllCores = 0 # MDS is unstable in parallel mode
# create a new 'Table To Points'
tableToPoints1 = TableToPoints(Input=tTKDimensionReduction2)
tableToPoints1.XColumn = "Component_0"
tableToPoints1.YColumn = "Component_1"
tableToPoints1.a2DPoints = 1
tableToPoints1.KeepAllDataArrays = 1
# create a new 'Mask Points' (to threshold on points)
maskPoints1 = MaskPoints(Input=tableToPoints1)
maskPoints1.OnRatio = 0
maskPoints1.GenerateVertices = 1
maskPoints1.SingleVertexPerCell = 1
# create a new 'Threshold'
threshold33 = Threshold(Input=maskPoints1)
threshold33.Scalars = ["POINTS", "treeID"]
threshold33.ThresholdMethod = "Between"
threshold33.LowerThreshold = 0.0
threshold33.UpperThreshold = 11.0
# create a new 'Threshold'
threshold34 = Threshold(Input=maskPoints1)
threshold34.Scalars = ["POINTS", "treeID"]
threshold34.ThresholdMethod = "Between"
threshold34.LowerThreshold = 12.0
threshold34.UpperThreshold = 14.0
# save the output
SaveData("MDS_trees.csv", threshold33)
SaveData("MDS_centroids.csv", threshold34)
|
To run the above Python script, go to your ttk-data directory and enter the following command:
pvpython python/mergeTreeClustering.py
- Isabel.cdb: a cinema database containing 12 regular grids.
Outputs
MDS_trees.vtu: the output points in 2D MDS (MultiDimensional Scaling) corresponding to the input trees. The 'ClusterAssignment' array contains the clustering assignments.MDS_centroids.vtu: the output points in 2D MDS (MultiDimensional Scaling) corresponding to the centroids.
C++/Python API
BlockAggregator
CinemaProductReader
CinemaReader
DimensionReduction
FlattenMultiBlock
MergeTree
MergeTreeClustering
MergeTreeDistanceMatrix
