Persistent Generators AT¶
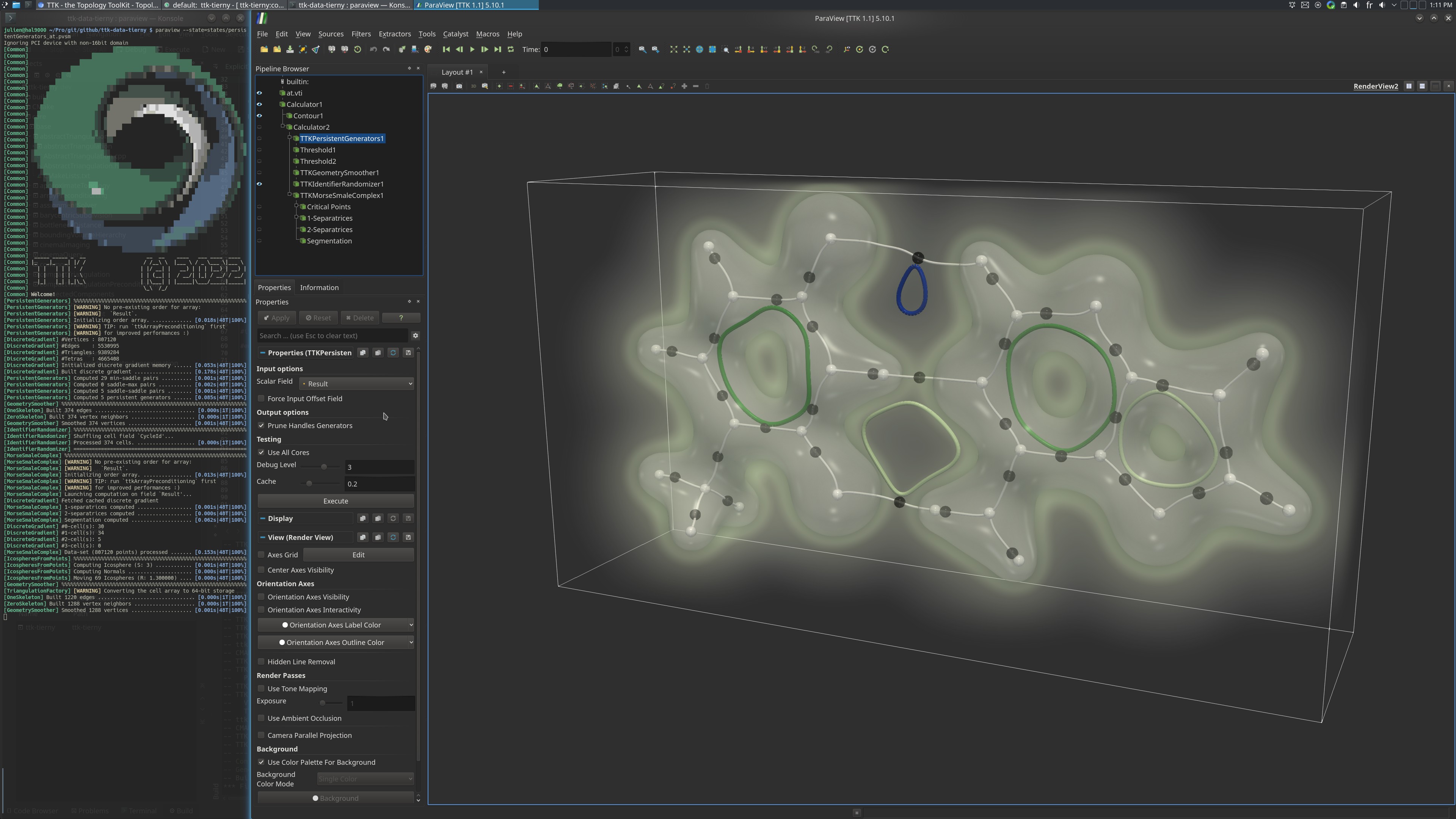
Pipeline description¶
This example computes the persistent one-dimensional generators of the electronic density of the Adenine-Thymine molecular complex. These generators characterize the cycles in the interactions between atoms, whether they are covalent (cycles in the carbonate chains), or weaker non-covalent interactions (cycles in the hydrogen bonds).
Generators are computed using PersistentGenerators. For context, the one-dimensional separatrices of the MorseSmale complex are computed using MorseSmaleComplex and shown in the ParaView example.
The python script simply computes the generators and saves the result as a .vtp file.
ParaView¶
To reproduce the above screenshot, go to your ttk-data directory and enter the following command:
paraview states/persistentGenerators_at.pvsm
Python code¶
1 2 3 4 5 6 7 8 9 10 11 12 | |
To run the above Python script, go to your ttk-data directory and enter the following command:
pvpython python/persistentGenerators_at.py
Inputs¶
- at.vti: A molecular dataset: a three-dimensional regular grid with 1 scalar field, the electronic density in the Adenine Thymine complex.
Outputs¶
PersistentGeneratorsAt.vtp: the raw one-dimensional generators of the field.